jMetalPy: Python version of the jMetal framework¶
Warning
Documentation is WIP!! Some information may be missing.
Contents:
Installation steps¶
Via pip:
$ pip install jmetalpy
Via Github:
$ git clone https://github.com/jMetal/jMetalPy.git
$ pip install -r requirements.txt
$ python setup.py install
Features¶
The current release of jMetalPy (v0.9.0) contains the following components:
- Algorithms: local search, genetic algorithm, evolution strategy, simulated annealing, random search, NSGA-II, SMPSO, OMOPSO, MOEA/D, GDE3. Preference articulation-based algorithms; G-NSGA-II and SMPSO/RP; Dynamic versions of NSGA-II and SMPSO.
- Parallel computing based on Apache Spark and Dask.
- Benchmark problems: ZDT1-6, DTLZ1-2, FDA, LZ09, unconstrained (Kursawe, Fonseca, Schaffer, Viennet2), constrained (Srinivas, Tanaka).
- Encodings: real, binary, permutations.
- Operators: selection (binary tournament, ranking and crowding distance, random, nary random, best solution), crossover (single-point, SBX), mutation (bit-blip, polynomial, uniform, random).
- Quality indicators: hypervolume, additive epsilon, GD, IGD.
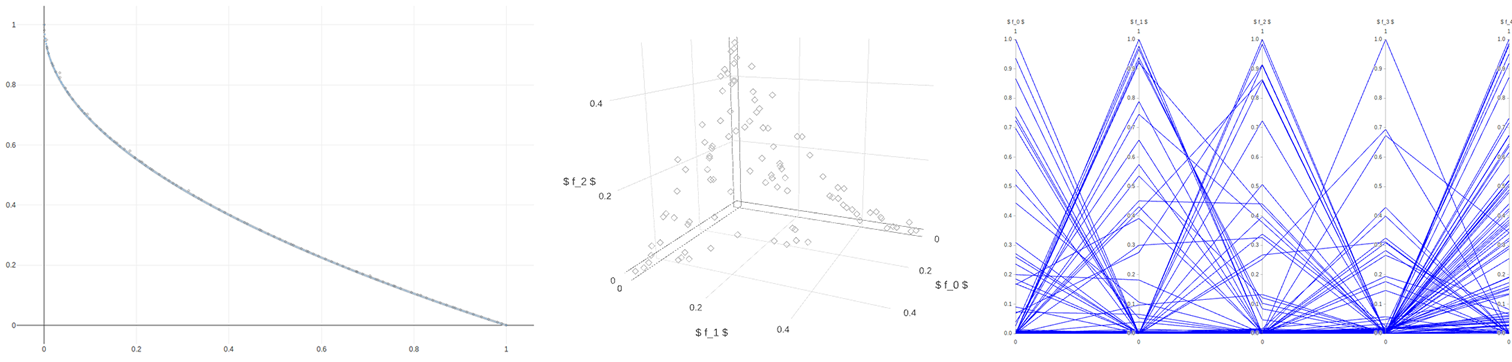
- Pareto front plotting for problems with two or more objectives (as scatter plot/parallel coordinates/chordplot) in real-time, static or interactive.
- Experiment class for performing studies either alone or alongside jMetal.
- Pairwise and multiple hypothesis testing for statistical analysis, including several frequentist and Bayesian testing methods, critical distance plots and posterior diagrams.